- C-MORE Home
- Member Area
- What is Microbial Oceanography?
- What is C-MORE?
- C-MORE Hale
- Research
- Theme I: Microbial Biodiversity
- Theme II: Metabolism and C, N, P & Energy Flow
- Theme III: Remote and Continuous Sensing; Links to Climate Variablilty
- Theme IV: Ecosystem Modeling, Simulation, and Prediction
- Research Cruises
- Data
- Education & Outreach
- People
- Publications
- Microscopy Images
- Professional Development Training Program
- Summer Course
- C-MORE Highlights
- Meeting Facilities
- Contact Us
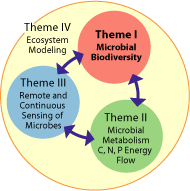 Research Themes
Research Themes
Theme I: Marine microbial biodiversity: from genomes and cultivation to ecology

Michael S. Rappé (Theme I Leader)
University of Hawai‘i at Manoa (UH)
C-MORE Theme I is focused on all aspects of microbial diversity found in marine plankton. Life has its origins in the sea, and its primordial form was microbial in nature. Given the deep and rich evolutionary history of microbial life, spanning > 3.5 billion years, the diversity of marine microbes is immense in scope. This diversity is expressed in all aspects of marine microbial life : organismal, genomic, physiological and ecological. C-MORE’s Theme I centers on cataloguing, categorizing, archiving and interpreting these many expressions of microbial diversity, in the specific contexts of the living ocean system. Theme I activities will therefore necessarily be tightly integrated with CMORE’s other central Themes.
Theme I will approach microbial diversity from two fundamentally different, yet allied and integrated perspectives :
- Cultivation-dependent approaches, whose aim is to isolate, maintain, and characterize the major microbial species inhabiting the waters of the North Pacific Subtropical Gyre. These cultivars will include representatives from diverse phyla of Bacteria and Archaea. The cultures will also span a broad spectrum of physiologies, including photoautotrophs, photoheterotrophs, chemolithoautotrophs, and chemoorganotrophs. In addition to cellular isolates, the viral predators of these key microbial players will also be isolated and characterized. One major goal is to discover and culture novel, ecologically relevant microorganisms, and to establish foundational model systems to better interpret microbial physiology, function and ecology.
 Cultivation independent approaches have dramatically altered our understanding of the nature and extent of microbial
diversity on planet Earth today. The number of uncultured microbes vastly outnumbers those now currently cultivated and characterized. Some of these
naturally occurring undomesticated microbes represent new microbial phyla, and probably new physiological diversity. Even without cultivars,
these new groups can now be better understood by tracking their genes and genomes in the environment. Consequently, Theme I will also employ
cultivation-independent approaches to identify specific microbial lineages, and quantify their spatial and temporal variability. Additionally,
newly developed environmental genomic approaches will allow characterization of whole microbial communities from genome-wide perspectives. Using
metagenomics, Theme I will explore the collective genomic repertoire of microbial communities sampled over time and space. These activities will
provide important data on the dynamic “parts list” of the collective microbial plankton. This will provide a starting point for identifying key
organisms for isolation, interpreting metabolic diversity (C-MORE Theme II), identifying targets for in situ sensing (C-MORE Theme III), and provide
data for more holistic modeling of complex microbial community dynamics and biogeochemical transformations (C-MORE Theme IV).
Cultivation independent approaches have dramatically altered our understanding of the nature and extent of microbial
diversity on planet Earth today. The number of uncultured microbes vastly outnumbers those now currently cultivated and characterized. Some of these
naturally occurring undomesticated microbes represent new microbial phyla, and probably new physiological diversity. Even without cultivars,
these new groups can now be better understood by tracking their genes and genomes in the environment. Consequently, Theme I will also employ
cultivation-independent approaches to identify specific microbial lineages, and quantify their spatial and temporal variability. Additionally,
newly developed environmental genomic approaches will allow characterization of whole microbial communities from genome-wide perspectives. Using
metagenomics, Theme I will explore the collective genomic repertoire of microbial communities sampled over time and space. These activities will
provide important data on the dynamic “parts list” of the collective microbial plankton. This will provide a starting point for identifying key
organisms for isolation, interpreting metabolic diversity (C-MORE Theme II), identifying targets for in situ sensing (C-MORE Theme III), and provide
data for more holistic modeling of complex microbial community dynamics and biogeochemical transformations (C-MORE Theme IV).
[ Top of Page ]



